Speaker
Description
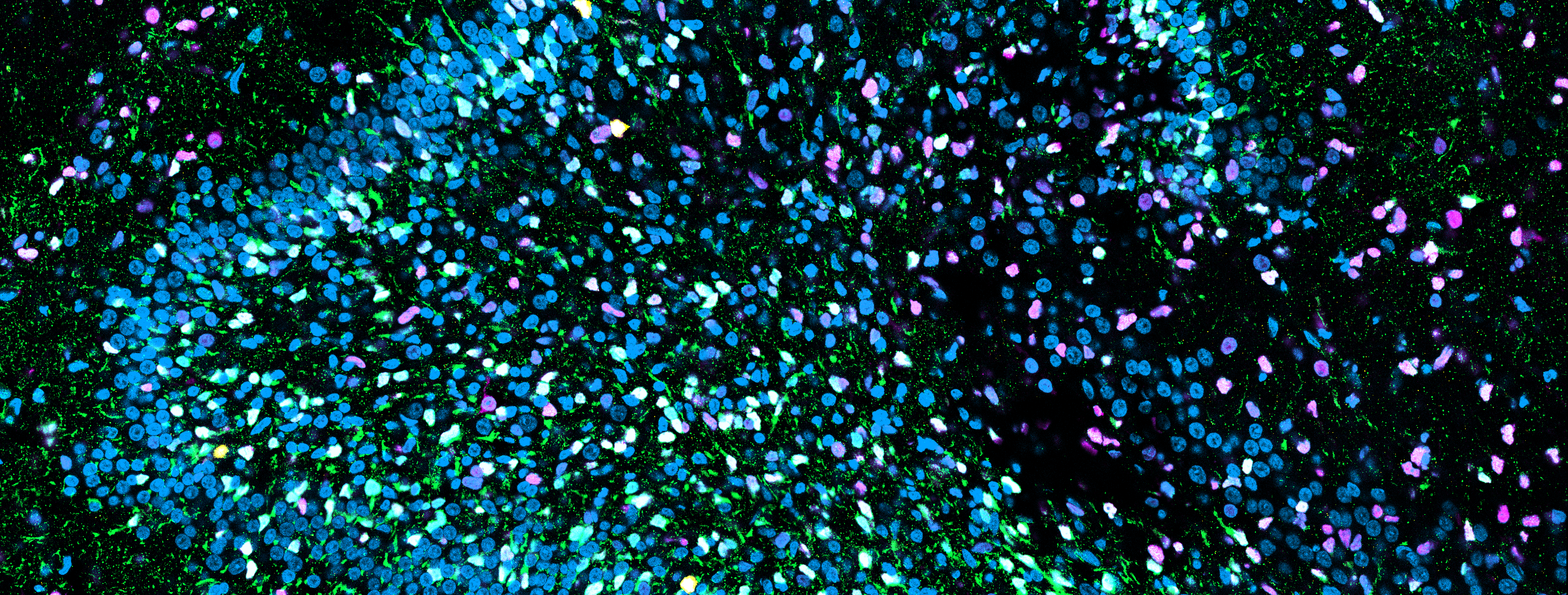
Spatial multi-omic technology has the potential to reveal the molecular foundations of neurodegeneration, uncovering both shared pathological responses and discrete signatures of vulnerability across clinically distinct neurodegenerative diseases. Using GeoMx spatial profiling, we quantified mRNA in CA1 pyramidal neurons with and without tau pathology in 6 primary age-related tauopathy (PART cases), 6 Alzheimer's disease (AD) cases, and 4 control cases. We found that tau pathology was the primary factor influencing transcriptional variation rather than disease classification. Differential gene expression analysis revealed an unexpected pattern: while synaptic genes were downregulated in disease neurons overall, tau-positive neurons showed upregulation of synaptic genes compared to tau-negative neurons in the same disease. Using unsupervised machine learning, we identified two distinct expression patterns associated with tau pathology that spanned both conditions. These transcriptional patterns were validated using transfer learning in an independent dataset of cortical neurons with tau pathology in AD, demonstrating that these changes are consistent across brain regions. Hybridization chain reaction studies in an expanded cohort confirmed the upregulation of select genes in tau-positive neurons.
Our findings highlight the power of molecular analysis stratified by pathology and suggest that the tau pathology in PART and AD may induce a shared molecular program despite occurring in different contexts. This has potential implications for therapeutic approaches targeting tau pathology in both conditions. We are currently developing an affordable, FFPE-compatible platform to spatially interrogate mRNA concurrently with a panel of known neuropathological markers. This expanded capability will allow us to correlate gene expression patterns with multiple protein markers simultaneously at subcellular resolution, providing a more comprehensive understanding of the cellular microenvironment in tauopathies and other neurodegenerative conditions.
| Author(s) | *Ryan Palaganas* 1, Chaichontat Sriworarat 1, Ernest M. Meyer 3, Javier Redding-Ochoa 4, Olga Pletnikova 4,5, Haidan Guo 4, William R. Bell 6, Juan C. Troncoso 4,7, Richard L. Huganir 1,2, Meaghan Morris 2,4,8, Genevieve L. Stein-O’Brien 1,2,7,8 |
|---|---|
| Affiliation(s) | "1. Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA, 2. Kavli Neuroscience Discovery Institute, Baltimore, MD 21218, USA 3. UPMC Hillman Cancer Center Cytometry Facility, University of Pittsburgh, Pittsburgh, PA 15232, USA, 4. Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA, 5. Department of Pathology and Anatomical Sciences, University at Buffalo, Buffalo, NY 14203, USA, 6. Department of Pathology and Laboratory Medicine, Indiana University School of Medicine, Indianapolis, IN 46202, USA, 7. Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA, 8. Co-corresponding authors" |