Speaker
Description
The human brain is a complex organ, and its features of neurodevelopment and disease remain largely unexplored due to limited accessibility of living tissue. Recently, the human induced pluripotent stem cells (hiPSC) technology allowed to model the human brain in vitro, generating 3D organoids.
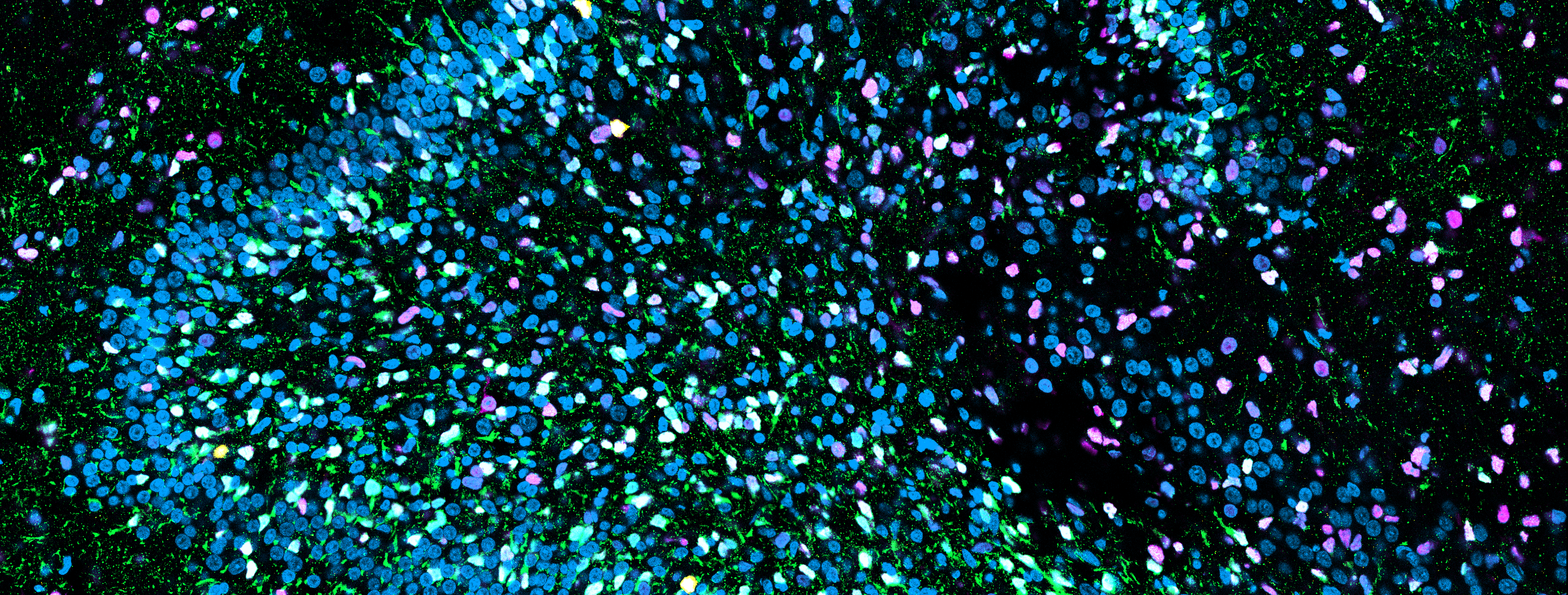
We developed brain and region-specific brainstem organoids, characterized them by fluorescent IHC to evaluate neuronal differentiation and brain regionalization, and bulk RNA sequencing followed by deconvolution to infer cell compositions and gene expressions. In parallel, label-free Raman images were collected on organoid slices for their non-invasive biomolecular characterization.
Since energy metabolism is a crucial element of neurodevelopment and can constitute a common pathway through different diseases, we performed untargeted metabolic signature analysis of developing brain organoids over time.
Comprehensively, the analyses performed on control brain organoids paved the way to model and characterize neurodevelopmental disease-derived organoids. They constitute an unvaluable asset in the study of neurodevelopmental disorders for which viable animal models are not available: in this context, we focused on the study of Congenital Central Hypoventilation Syndrome (CCHS). CCHS is a rare genetic disorder affecting the autonomic nervous system (ANS) and central chemosensitivity caused by the loss of retrotrapezoid nucleus (RTN) neurons in the brainstem. Mutations of the PHOX2B master gene affect the development of the ANS and central structures (RTN) that participate in breathing control. Given its fundamental role, CCHS animal models die in utero: to overcome this issue and have an insight in PHOX2B’s function during the first stages of human neurodevelopment, we used CCHS patient-derived iPSCs carrying different PHOX2B mutations to generate brain and brainstem organoids that contain cytoarchitectures resembling central chemoreceptors.
This new personalized disease-in-a-dish model of CCHS will allow to identify molecular and cellular defects induced by PHOX2B mutations as well as modelling for drug discovery/screening for therapeutic perspectives.
| Author(s) | Eleonora PISCITELLI*(a,f), Simona DI LASCIO(b), Filippo CHIESA(b), Alice CHIODI(a), Cinzia COCOLA(a), Paride PELUCCHI(a), Daniela GAGLIO(c), Marcella BONANOMI(c), Renzo VANNA(d), Victor ALCOLEA RODRIGUEZ(d), Ana Lucia CUADROS GAMBOA(b), Martina BERTOCCHI(b), Diego FORNASARI(b), Roberta BENFANTE(b,e,f) |
|---|---|
| Affiliation(s) | "a) CNR – Istituto di tecnologie biomediche (CNR-ITB), Segrate, Italy, b) Dept BIOMETRA, Università degli Studi di Milano, Milan, Italy, c) CNR-Istituto di Bioimmagini e Sistemi Biologici Complessi (CNR-IBSBC), Segrate, Italy, d) CNR - Istituto di Fotonica e Nanotecnologie (CNR-IFN), P.zza Leonardo da Vinci 32, 20133, Milan, Italy, e) CNR - Institute of Neuroscience (CNR-IN), Vedano al Lambro (MB), Italy, f) NeuroMi - Milan Center for Neuroscience, University of Milano Bicocca, Milan, Italy" |